Simultaneous identification of antigen-specific B and T cell clones using dCODE Dextramer®.
The immune system is a complex and dynamic landscape of cellular and biomolecular interactions. In the past, the characterization of immune responses has been limited in scope, often focused on a single cell type or handful of biomolecules. However, with the advent of today’s technologies, we can now examine the diversity of immune cells and their behavior with unparalleled resolution, revealing a world of intricate interactions.
While technologies for broader immune cell detection have been in use for several years, the race to understand the variation in COVID-19 progression and elucidate response mechanisms to novel mRNA-based vaccines cast light on the need to access information about multiple immune cell types.
Various studies discuss the role of T cells and B cells in subjects naturally infected with SARS-CoV-2 or responding to vaccination.1-4 All highlight with increasing detail the coordinated interplay between enriched B cells, activated CD4+ T cells, and expanded effector CD8+ T cells,² plus differences in cellular responses between infected and vaccinated individuals.³
Characterizing both cell populations can also help better understand other indication areas. For example, a concurrent examination can open new avenues for cancer vaccines. Known to contribute to anti-tumor mechanisms as immune regulators, antigen-presenting cells,⁴ and hubs of humoral immunity,⁵ B cells may magnify or lengthen T cell-mediated cytotoxicity.
Similarly, examining the interplay between T and B cells in the same sample can deliver direct evidence of therapy effects. For example, Bi-specific Autoantigen-T cell engagers (BiAATEs) are being explored to target subsets of autoreactive B cells in the treatment of autoimmune diseases.⁶ The BiAATEs induce T cell-dependent depletion of very specific B cells. Analyzing the cytotoxic cells and their target cells in the same sample provides insights into the therapy’s mode of action.
Two practical aspects came to light in studies of immune cell populations during COVID-19 progression. First, adequate reagents to identify antigen-specific cells are essential.⁷ Second, analyzing both B and T cells at the same time and in the same sample circumvents the depletion of specimens that are often difficult to acquire.¹ Mindful of these two aspects, here we discuss the simultaneous, single-cell multi-omic characterization of B and T cells using Immudex’s dCODE® reagents.
dCODE Dextramer® and dCODE Klickmer® reagents are DNA barcoded multimers that allow you to distinguish immune cells of different specificities. dCODE Dextramer® reagents feature MHC-peptide complexes to detect T cells (MHC I for CD8+ T cells or MHC II for CD4+ T cells), whereas dCODE Klickmer® reagents are loaded with any biotinylated molecule to serve as antigen, allowing you to, for example, detect antigen-specific B cells.
Simultaneous detection of antigen-specific T and B cells using barcoded dCODE® reagents
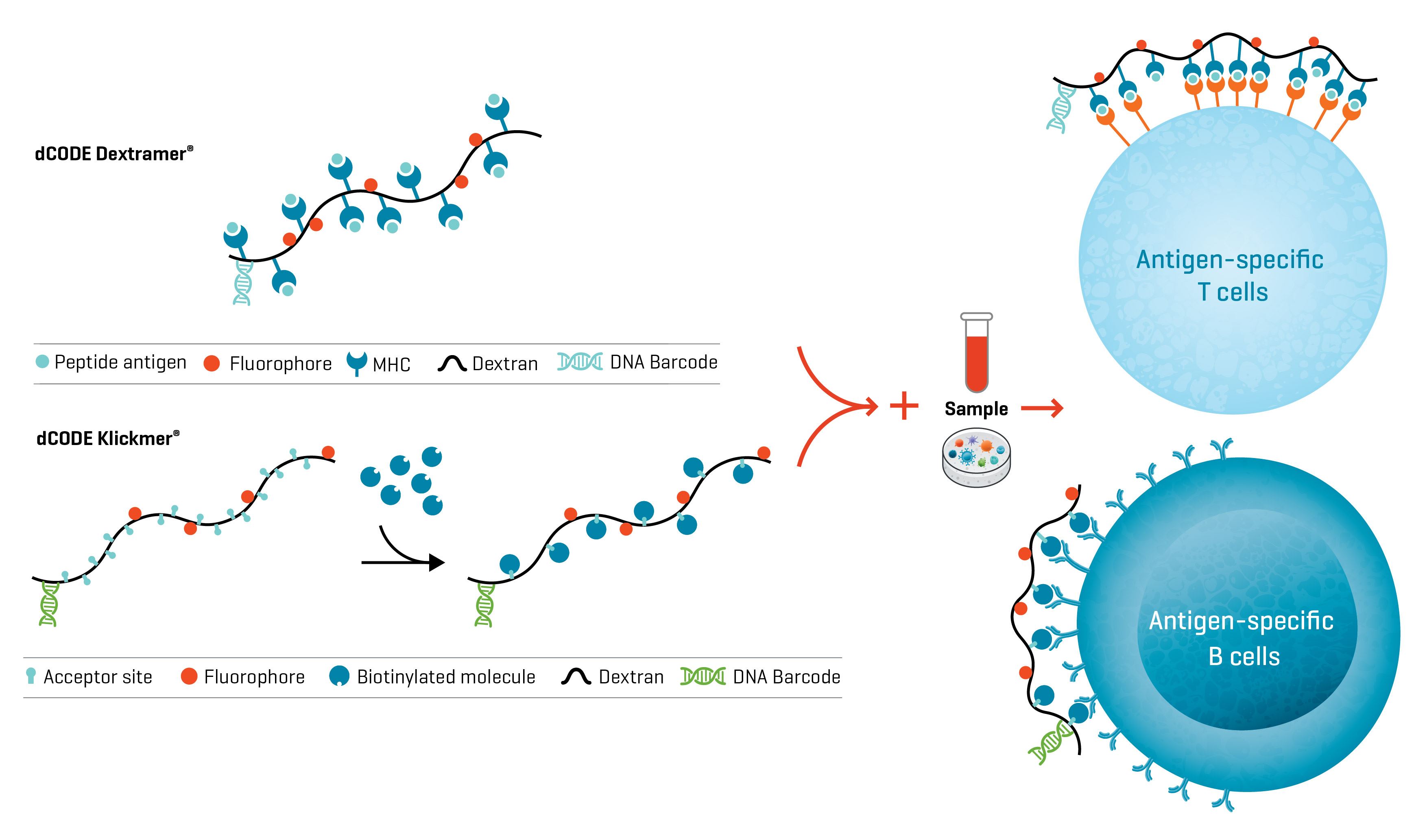
Fig. 1. d CODE® reagents from Immudex allow the detection and analysis of multiple antigen-specific population subsets of T cells and B cells in a single sample.
Combining the unique barcoding of these reagents with a single-cell analysis platform like the 10x Chromium Single-Cell Analysis System or the BD Rhapsody™ Single-Cell Analysis System lets you simultaneously evaluate T and B cell kinetics in response to multiple antigens. You can examine BCR and TCR clonotypes, gene expression, and surface marker profiles of both cell types at single-cell resolution.
The general workflow for this multiplexed analysis is straightforward:
Workflow for the simultaneous single-cell analysis of antigen-specific B and T cells
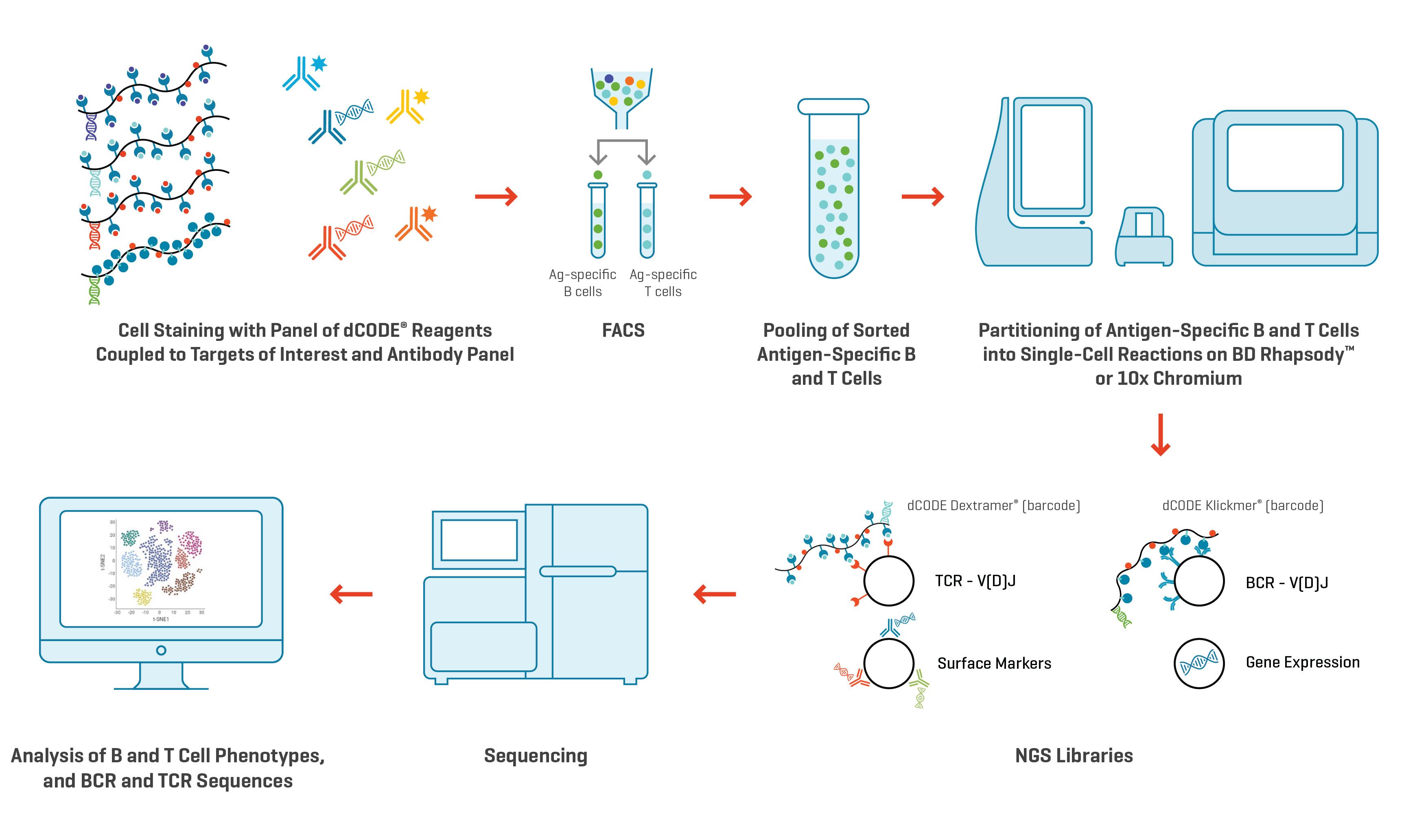
Fig. 2. Workflow to examine clonotypes of antigen-specific B cells and T cells using multiplexed dCODE® reagents on the 10x Chromium Single-Cell Analysis System or the BD Rhapsody™ Single-Cell Analysis System.
To demonstrate the ease and informational output of simultaneously examining T and B cells, we identified TCR and BCR clonotypes in blood samples taken from a COVID-vaccinated individual (HLA type: HLA-A*0101; HLA-B*0702). An initial sample was taken immediately after recovery from a breakthrough SARS-CoV-2 infection, and a second blood draw was done five weeks later.
Our dCODE Dextramer® panel to detect and identify antigen-specific T cells covered eight SARS-CoV-2 epitopes, three CMV epitopes, three EBV epitopes, and one influenza epitope (Table 1). This panel was combined with a set of dCODE Klickmer® reagents to detect spike protein-specific B cells. We recommend dual staining of B cells with two fluorophores to reduce background. In this case, only one reagent was DNA barcoded, but dCODE Klickmer® reagents with various fluorophores are available upon request.
Samples were incubated with the panel reagents for 20 minutes. In a second incubation (20 minutes), the dCODE® reagent-labeled cells were stained with antibodies to allow FACS sorting based on the detection of a viability marker, as well as Klickmer® and Dextramer® reagents, CD3, CD8, CD14, and CD19. Totalseq C antibodies representing canonical immune cell markers were also included (Table 2). After rinsing, the cells were sorted according to the gating strategy depicted in Figure 3. Approximately 50–60,000 antigen-specific B cells and 70,000 antigen-specific T cells were sorted for each time point.
Table 1. Panel of dCODE Dextramer® reagents used to detect antigen-specific T cells
Cat No. |
Allele |
Peptide |
Antigen |
Organism |
Fluorochrome |
DNA barcode |
|
WA05846dXG |
A*0101 |
LTDEMIAQY |
S |
SARS-CoV-2 |
PE |
A |
|
WA05847dXG |
A*0101 |
WTAGAAAYY |
S |
SARS-CoV-2 |
PE |
B |
|
WH05879dXG |
B*0702 |
SPRRARSVA |
S |
SARS-CoV-2 |
PE |
C |
|
WH06103dXG |
B*0702 |
APHGVVFL |
S |
SARS-CoV-2 |
PE |
D |
|
WA05973dXG |
A*0101 |
FTSDYYQLY |
ORF3a |
SARS-CoV-2 |
PE |
E |
|
WA05972dXG |
A*0101 |
TTDPSFLGRY |
ORF1ab |
SARS-CoV-2 |
PE |
F |
|
WH06032dXG |
B*0702 |
IPRRNVATL |
ORF1ab |
SARS-CoV-2 |
PE |
H |
|
WH05842dXG |
B*0702 |
SPRWYFYYL |
N |
SARS-CoV-2 |
PE |
I |
|
WA03579dXG |
A*0101 |
Nonsense peptide |
Negative control |
PE |
J |
|
|
WB02666dXG |
A*0201 |
Nonsense peptide |
Negative control |
PE |
K |
|
|
WI03233dXG |
B*0801 |
Nonsense peptide |
Negative control |
PE |
L |
|
|
WA02131dXG |
A*0101 |
VTEHDTLLY |
pp50 |
CMV |
PE |
M |
|
WA03410dXG |
A*0101 |
CTELKLSDY |
NP |
Influenza |
PE |
N |
|
WH02136dXG |
B*0702 |
TPRVTGGGAM |
pp65 |
CMV |
PE |
O |
|
WH02135dXG |
B*0702 |
RPHERNGFTVL |
pp65 |
CMV |
PE |
P |
|
WH03287dXG |
B*0702 |
RPQGGSRPEFVKL |
BMRF1 |
EBV |
PE |
Q |
|
WH02166dXG |
B*0702 |
RPPIFIRRL |
EBNA 3A |
EBV |
PE |
R |
|
WH02165dXG |
B*0702 |
QPRAPIRPI |
EBNA 6 |
EBV |
PE |
S |
Table 2. Antibodies used to characterize single cells according to canonical immune cell markers
Totalseq C antibodies |
BioLegend Article No. |
|
Totalseq C0384 anti IgD (Ig class switching) |
348245 |
|
Totalseq C0046 anti CD8 (T cell pan marker) |
344753 |
|
Totalseq C0154 anti CD27 (memory marker of B cells) |
302853 |
|
Totalseq C0051 anti CD14 (monocytes) |
367137 |
|
Totalseq C0034 anti CD3 (T cell pan marker) |
300479 |
|
Totalseq C0050 anti CD19 (B cell pan marker) |
302265 |
Gating strategy to sort antigen-specific T and B cells from a single sample
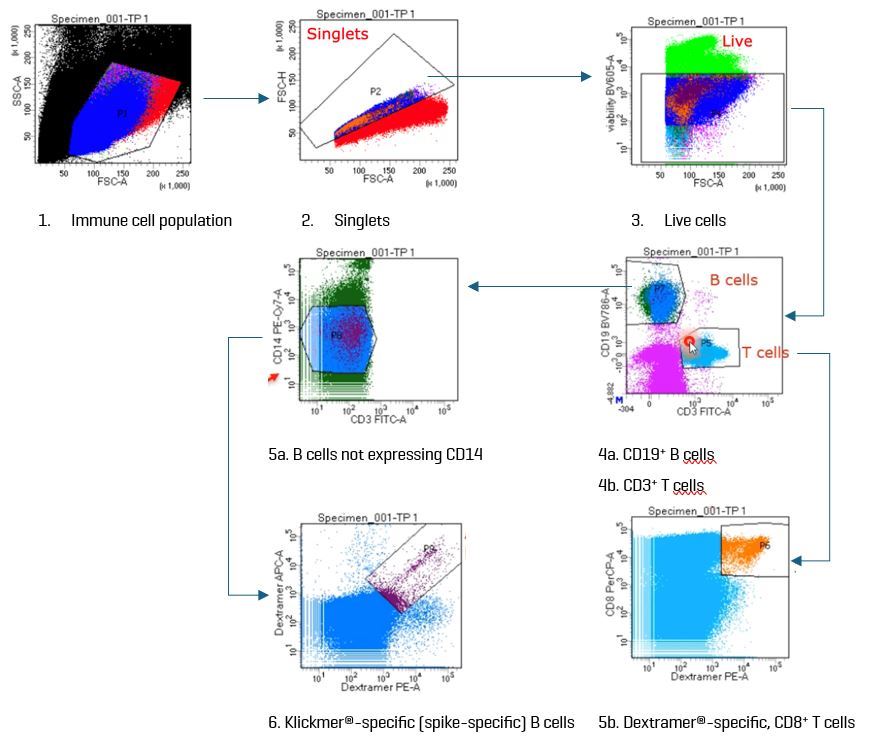
Fig. 3. The gat ing strategy used in this study first sorted live, single cells into B and T cells based on the expression of CD19 and CD3, respectively. In a subsequent gate, spike-specific B cells were captured as CD14–Klickmer®-positive B cells. Captured T cells were CD8+.
We then pooled the sorted antigen-specific T and B cells and loaded them onto the Chromium platform from 10x Genomics for singularization, cDNA amplification, and generation of libraries for Illumina sequencing of antibody barcodes, gene expression, and V(D)J clonotype. We processed the resulting FASTQ data files with the Cell Ranger Multi Pipeline.

Figure 4. t-SNE plot of all T and B cells captured in the single-cell analysis. B cells captured at the first time point (immediately after recovery) are shown as blue dots and at the second time point (5 weeks post-recovery) as orange dots. Non-B cells are shown as gray dots.
Spike-specific B cells expanded between the first sample collection immediately after recovery from COVID-19 and the second sample taken five weeks later (114 at time point 1 vs 387 at time point 2). The expanded B cells exhibited upregulated CD27 expression and downregulated IgD, indicating a memory phenotype. Compared to all other B cells, the following genes were upregulated in the spike-specific B cells: CRIP1, IFI30, PLAC8, SUB1, S100A10, and CIB1.

Figure 5. Among the most frequent BCR clonotypes sequenced, six were more prevalent at time point 2 (5 weeks post recovery; orange) compared to time point 1 (immediately after recovery; blue). However, none of these clonotypes mapped to the spike-specific B cells that were captured.
The overall number of B cells captured at both time points were similar (~4700 cells) and the population also exhibited high BCR clonotype diversity (>4400 clonotypes) at both time points. Six of the ten most represented clonotypes were found in greater proportion at time point 2, although none of these clonotypes mapped to the spike-specific B cells that we captured. The analysis also provided CDR3 sequences of the immunoglobulin heavy and kappa chains (data not shown).

Figure 6. t-SNE plot of all T and B cells captured in the single-cell analysis. T cells captured at the first time point (immediately after recovery) are shown as red dots and at the second time point (5 weeks post-recovery) as green dots.
Looking at the antigen-specific T cells captured in our analysis revealed that a large proportion of the CD8+ T cell response (ie., number of cells) was directed toward epitope TTDPSFLGRY of the open reading frame polyprotein ORF1ab. This coronavirus protein has been described as an immunodominant epitope of SARS-CoV-2.⁸ CD8+ T cells targeting this epitope increased significantly in number between time point 1 (red dots) and time point 2 (green dots; >2000 identified cells). ORF1ab-specific CD8+ T cells showed upregulated expression of STMN1, TUBA1B, HIST1H4C, HMGB2, TUBB, H2AFZ, and LGALS1 compared to all other CD8+ T cells.


Figure 7. Among the most frequent TCR clonotypes sequenced, two clonotypes (2 and 7) recognized the immunodominant ORF1ab epitope and were expanded at time point 2 (5 weeks post recovery; orange) compared to time point 1 (immediately after recovery; blue).
The overall population of captured CD8+ T cells nearly doubled at time point 2 compared to time point 1 (5659 vs. 3382). The TCR clonotype repertoire of the population was less diverse than the BCR clonotype repertoire of the B-cell population. However, among the top 10 most frequent clonotypes, two recognized the ORF1ab epitope mentioned above. These two clonotypes were also more prevalent at time point 2 compared to time point 1. CDR3 sequences of the top clonotypes were also obtained (data not shown).
In summary, data from the simultaneous analysis of antigen-specific T and B cells in this sample show:
Watch the on-demand webinar for a more detailed description of this case study.
The case study above demonstrates how examining two pivotal cell types within the same sample generates multidimensional details about the immune response that can be leveraged for hypothesis generation. As a tool, using dCODE® reagents in combination with a single-cell analysis platform allows us to delve deeper into the pathogenesis of infections and potentially discover novel treatment strategies.
Interested in looking closely at other immune cells?
We have also demonstrated in this scientific poster that the simultaneous characterization of immune cells at single-cell resolution can be expanded beyond B cells and CD8+ T cells to include CD4+ T cells, invariant natural killer (iNK) cells, and mucosal-associated invariant T (MAIT) cells. That’s five cell types in one sample.
So, go ahead and unlock the crucial details of complex immune responses by characterizing multiple immune cell types at high resolution using flexible dCODE Dextramer® and cCODE Klickmer® reagents.
Contact us to learn how.
¹Newell, K. L. et al. 2022. Simultaneous analysis of antigen-specific B and T cells after SARS-CoV-2 infection and vaccination. Cytometry 101: 474. 10.1002/cyto.a.24563
² Sureshchandra, S. et al. 2021. Single-cell profiling of T and B cell repertoires following SARS-CoV-2 mRNA vaccine. JCI Insight 6: e153201. 10.1172/jci.insight.153201
³ Lo Tartaro, D. et al. 2023. Detailed characterization of SARS-CoV-2 specific T and B cells after infection or heterologous vaccination. Front. Immunol. 14: 1123724. 10.3389/fimmu.2023.1123724
⁴ Wennhold, K. et al. 2019. B Cell-Based Cancer Immunotherapy. Transfus Med Hemother. 46: 36. 10.1159/000496166
⁵ Kaumaya, P.T.P. 2020. B-Cell Epitope Peptide Cancer Vaccines: A New Paradigm for Combination Immunotherapies with Novel Checkpoint Peptide Vaccine. Future Oncol. 16: 1767. 10.2217/fon-2020-0224
⁶ Perico, L. et al. 2024. Bi-specific autoantigen-T cell engagers as targeted immunotherapy for autoreactive B cell depletion in autoimmune diseases. Front. Immunol. 15: 1335998. 10.3389/fimmu.2024.1335998
⁷ De Baisi, S. et al. 2023. Analysis of antigen-specific T and B cells for monitoring immune protection against SARS-CoV-2. Curr. Protoc. 3: e636. 10.1002/cpz1.636
⁸ Gangaev, A. et al. Identification and characterization of SARS-CoV-2 specific CD8+ T cell response with immunodominant features. Nature Comm. 12: 2593. 10.1038/s41467-021-22811-y
Simultaneous identification of antigen-specific B and T cell clones using dCODE Dextramer®.
Simultaneous characterization of antigen-specific CD8+ and CD4+ T cells, B cells, iNKT cells and MAIT cells.
Fill in the form and one of our dedicated specialists will get in touch with you shortly.