Get the case study as a PDF.
Identification of Epitopes that Pose as Attractive Targets for Vaccination and Other T-Cell Therapies with Klickmer®
Background
CD4+T cells orchestrate protection from severe influenza. However, knowledge of epitopes and the molecular patterns associated with recognition across the population is lacking.
In this study, researchers identified several influenza epitopes from internal virus proteins using customized Klickmer® and apply this knowledge to explore the biochemical features that underpin CD4+ T-cell responses to influenza.
Study Description
Human peptide-HLA monomers were biotinylated and multimerized onto the Klickmer® backbone. The peptide-HLA Klickmer® reagents were then used for identification of HLA-restricted epitopes within three internal Influenza A Viral proteins, including M1, NP, and PB-1 in peptide expanded cell cultures from five HLA-DR1+ donors.
Results
To quantify and compare recognition of selected epitopes, peptide-HLA Klickmer® reagents were used to identify epitope-specific responses in five HLA-DR1+ donors (Fig. 1A).
Robust epitope-specific responses were detected in all donors to the control epitope HA306-318-PKY, with natural donor-specific variation in response magnitude (range, 6.9%–26.2% CD4+). Of the five internal epitopes tested, 24 out of 25 possible responses were positive for Klickmer® staining. Positive responses were defined as Klickmer® staining/total CD4+ T cells 3 100 > 0.5%; donor-4 DPF was negative).
The largest responses, whether measured in terms of size of CD4+ T-cell expansion or of the median fluorescence intensity (MFI) of Klickmer® positive cells, were consistently to M1129-142-GLI followed closely by HA306-318-PKY (Fig. 1B and 1C)
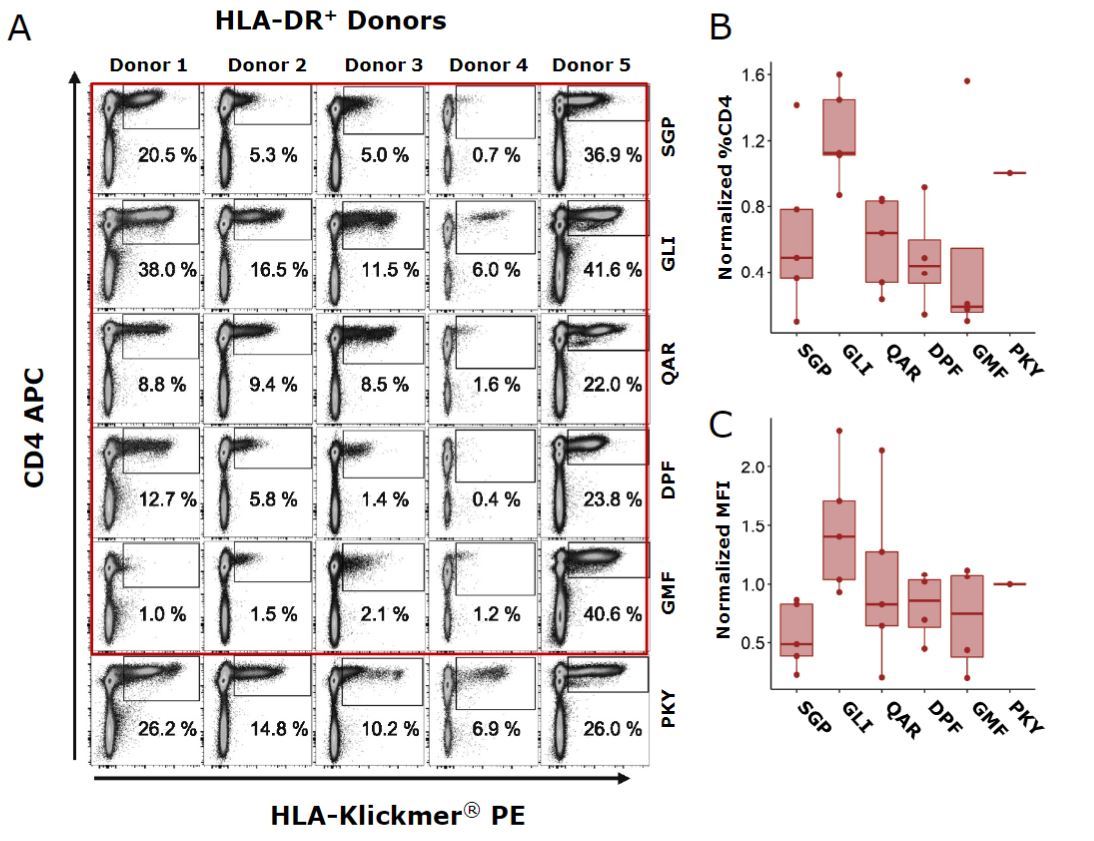
Fig. 1: Epitope recognition. A) Staining with peptide-HLA Klickmer®in five HLA-DR1+ donors for identification of epitope responses. Responses were measured as B) CD4+ T cell expansion or C) median fluorescence intensity
Conclusions
- HLA Klickmer® was successfully applied to characterize CD4+T-cell responses to influenza A virus
- Five internal epitopes commonly recognized by CD4+T cells in five HLA-DR1+subjects correlated with protective immunity were identified

