Get the case study as a PDF.
dCODE Dextramer® Reagents Identify Antigen-Specific Populations and Their TCR Clonotypes at Single-Cell Level
Adapted from Jacobsen, K. et. al. SITC 2019 poster P186
Background
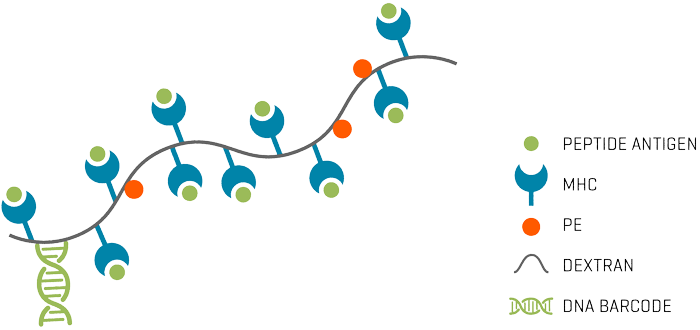
dCODE Dextramer® (10x) reagents are DNA barcoded MHC Dextramer® reagents designed for use with 10x Genomics Feature Barcode protocol for Single-Cell Immune Profiling (Fig. 1). Each unique barcode is specific for the MHC-peptide displayed on the dCODE Dextramer®.
Study Description
Goal: Detect antigen-specific T cells and their cognate T-cell receptors sequences in a human PBMC sample using a highly multiplexed panel of dCODE Dextramer® reagents.
- Healthy donor PBMC sample was stained with a pool of 50 different MHC I dCODE Dextramer® reagents displaying different viral and cancer epitopes
- dCODE Dextramer® positive cells were sorted by flow cytometry and loaded onto 10X Chromium controller
- Generation of three DNA libraries: 1) dCODE Dextramer® binders; 2) V(D)J sequences; 3) RNA expression. Each library sequenced by next-generation sequencing (Illumina)
Results
Four antigen-specific T-cell populations were identified: one for influenza (Flu), two for Epstein-Barr virus (EBV-1 and EBV-2), one for MART-1 (Fig. 2). For each population, the paired clonal TCR sequences were directly obtained and quantified.
Multiple TCR clones were identified for each antigen antigen-specific T-cell population (Table 1). Highly expanded TCR clones were found in viral T-cell populations (EBV-1 and EBV-2). Fewer expanded TCR clones were found for MART-1.
To confirm whether the identified MART-1-specific T-cells represent a naïve population, it is possible to interrogate the available gene expression profile.
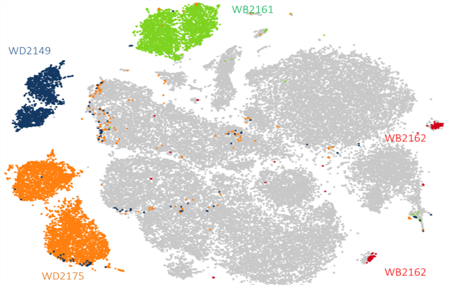
Fig. 2. Four antigen-specific T-cell populations identified after analysis of sequencing data
| Specificity (MHC-peptide) | Flu | EBV-1 | EBV-2 | MART-1 |
| Positive cells (number) | 2594 | 1846 | 4472 | 187 |
| Specific clones (number) | 732 | 166 | 389 | 180 |
| Most represented clone (number of cells) | 110 | 1241 | 2373 | 8 |
| Most represented clone (frequency) | 4% | 67% | 53% | 4% |
| Clone with >1 cell (number) | 251 | 19 | 11 | 1 |
| Clone with 1 cell (number) | 481 | 147 | 345 | 179 |
Table 1. TCR clones for each antigen-specific population
Conclusions
- dCODE Dextramer® technology enables the generation of highly multiplexed antigen-specificity data in a single experiment
- Combining dCODE Dextramer® and 10x Chromium represents a powerful tool for deep phenotyping of immune relevant cells
- Personalized dCODE Dextramer® libraries allow profiling of patients’ T cells and give a new understanding of T-cell immunity

