Get the case study as a PDF.
Identifying Shared Tumor Epitopes from Endogenous Retroviruses for High-Avidity Cytotoxic TILs using MHC Dextramer®
Bonaventure, P. et al. Identification of shared tumorepitopes from endogenous retroviruses inducing high-avidity cytotoxic T cells for cancer immunotherapy. Science Advances; 8(4). https://doi.org/10.1126/sciadv.abj3671
Background
Human endogenous retroviruses (HERVs) represent 8% of the human genome and may represent tumor antigens relevant for cancer immunotherapy. Bonaventure et al. developed a bioinformatic approach to identify shared CD8+ T cell epitopes derived from cancer-associated HERVs in solid tumors. Six candidates among the most commonly shared HLA-A2 epitopes (P1-6) with evidence of translation were selected for enrichment and immunological evaluation of epitope-specific CD8+ tumor-infiltrating leukocytes (TILs) from HLA-A2 patients with breast cancer using MHC Dextramer®.
Later on, the HERV-specific T cells were found to lyse patient-derived organoids. These shared virus-like epitopes may be of major interest in cancer vaccine or S DT TcelUlD-YbaseEdS iCmRImPTunoIONtherapy development, especially in tumors with low or intermediate mutational burden.
Study Description
MHC Dextramer® staining was performed on PBMCs after a 12-day culture (priming protocol) or on TILs expanded for 14 days after tumor dilaceration. Cells were washed in 2 ml of washing buffer (phosphate-buffered saline + 2% FBS + 2 mM EDTA) and stained for 10 min with MHC Dextramer® at room temperature before viability and surface marker staining. Washing was performed two times to avoid nonspecific MHC Dextramer® staining. CMV pp65 NLVPMVATV was used as a positive control and a non-natural irrelevant peptide (ALIAPVHAV) was used as a negative control.
Results
To test whether an adaptive immune response against HERV may exist in patients with cancer, we assessed Rby ESMUHLCT SDextramer® staining the presence of HERV epitope–specific T cells among polyclonally expanded TILs from HLA-A2 patients with triple-negative breast cancer without any peptide-specific stimulation. HERV-specific TILs were observed for at least one epitope in 7 of the 11 analyzed tumor samples, with variations in terms of epitope specificity and frequency from one patient to another. Epitopes P1, P4, and P6 were the most frequently recognized peptides, with an MHC Dextramer®-based identification in 4 of 11, 4 of 11, and 5 of 11 cases, respectively, whereas no significant staining was seen for P3 (Fig. 1).
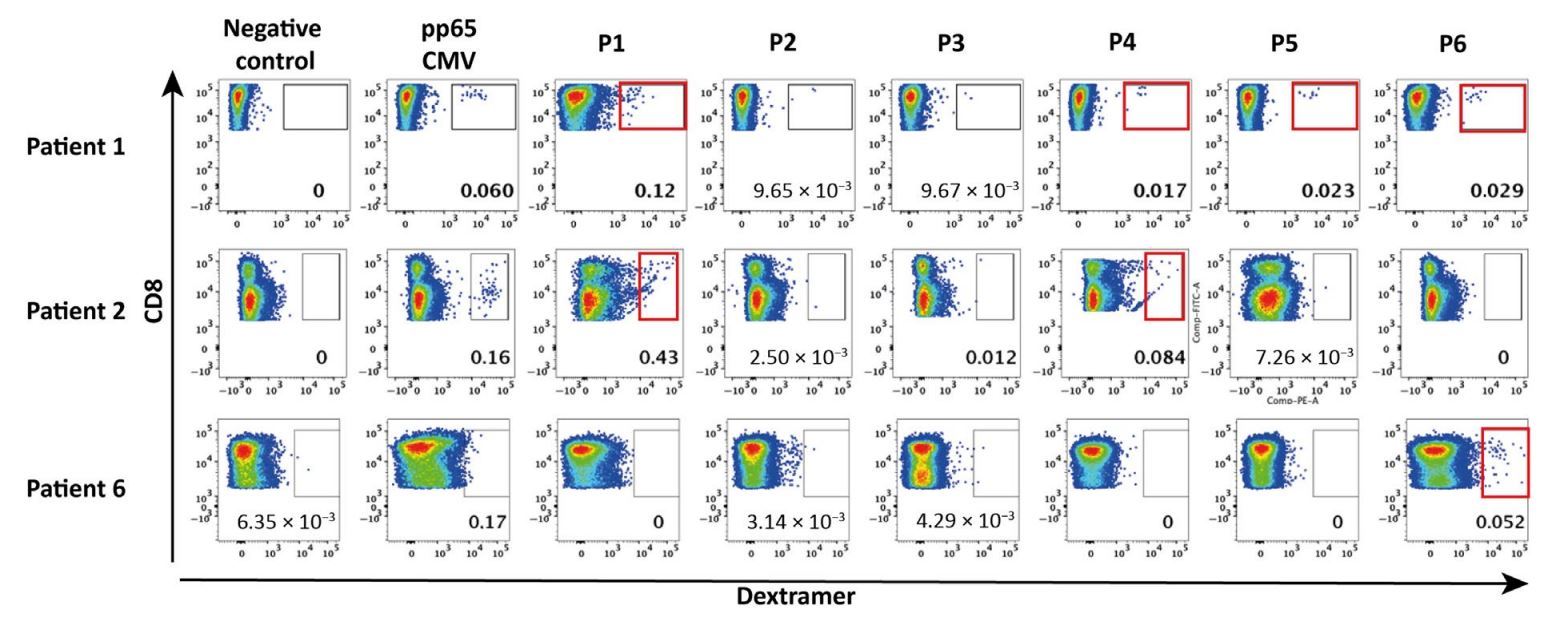
Fig.1: HERV-specific T cells are present among tumor-infiltrating T cells. Representative panels of dextramer staining for the six HERV epitopes in TILs from three HLA-2–positive patients with TNBC. Dextramer-positive cells were gated on CD8+ T cells. Negative control: Dextramer complexed to a non-natural irrelevant peptide (ALIAPVHAV)
Conclusions
- MHC Dextramer® efficiently identified all T cells populations specific for the selected epitopes that had been predicted to be shared among cancer patients
- Based on MHC Dextramer® staining, a higher prevalence of CD8+ T cells specific for HERV epitopes P1, P4, and P6 were observed in patients with triple-negative breast cancer

