Get in Touch with our Immune Monitoring Experts
Rigorous identification and validation of novel epitopes for today’s T-cell therapies
- Go multiplex: Generate highly multiplexed antigen mapping data for rapid epitope discovery and neoantigen screening in a single experiment using DNA barcoded dCODE Dextramer® reagents.
- High avidity: Dextramer® technology ensures that even low-affinity pMHC-TCR pairs are reliably detected, so you don’t miss potential candidates.
- Broad allele selection: From MHC Monomers to dCODE Dextramer® reagents, we offer the broadest selection of MHC I and MHC II alleles.
- Setup flexibility: Choose ready-to-use dCODE Dextramer® reagents, or build your own dCODE Dextramer® library with our peptide-receptive MHC Monomers and U-Load dCODE Dextramer® reagents.
- Validate and characterize: Validate epitopes by flow cytometry using MHC Dextramer® reagents, or link antigen specificity to gene expression, surface protein, and V(D)J sequences with a single-cell multi-omics dCODE Dextramer® approach.
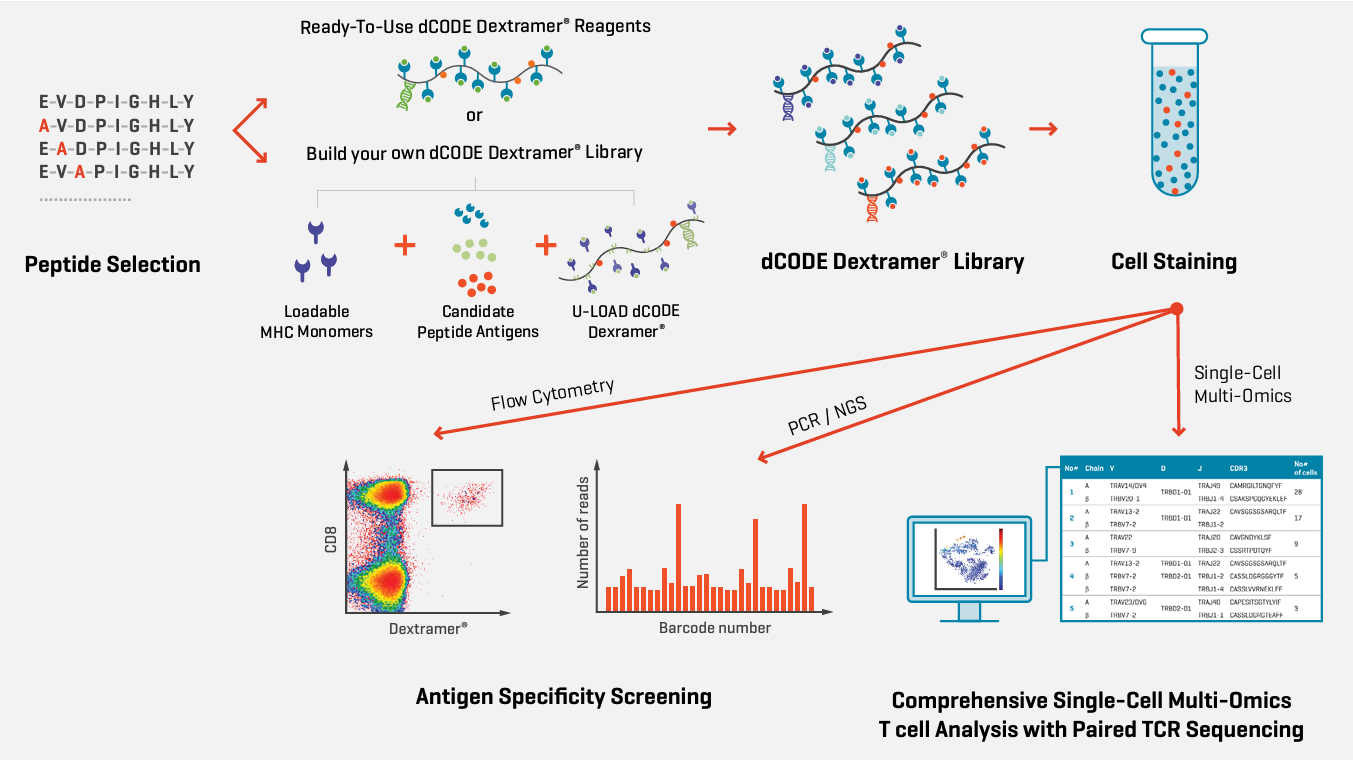
Multiplex screening of large epitope panels using dCODE Dextramer®. dCODE Dextramer® library is created using ready-to-use dCODE Dextramer® reagents or alternatively by building your own library using U-Load dCODE Dextramer® and loadable* MHC Monomers incubated with the candidate peptides. After staining the cell sample with the dCODE Dextramer® library, FACS-sorting is used to isolate the MHC-multimer-binding cells based on PE fluorescence intensity. The antigen-specificities are then identified by sequencing the DNA barcodes. Positive populations are validated by flow cytometry. TCR discovery can be performed at scale using libraries of dCODE Dextramer® reagents40, enabling up to 1000 different specificities to be screened in the same sample. The advantage of this single-cell dCODE Dextramer® approach is that it enables you to simultaneously identify the TCR sequences as well as the epitopes with which the TCRs interact.
Developing a novel TCR T cell therapy begins with screening epitopes that bind strongly to a relevant MHC allele and elicit the desired therapeutic effect. Our approach utilizes a library of DNA-barcoded dCODE Dextramer® reagents for large-scale profiling of epitope recognition by T cells. This method allows you to quantify binding TCR clonotypes, to identify which epitopes are being seen by T cells.
For deeper characterization, this approach can be combined with single-cell multi-omics to examine T-cell differentiation and activation markers, and evaluate immunogenicity. Epitopes can be further validated using the same Dextramer® technology, by flow cytometry. Every assay benefits from the high avidity of Dextramer® Technology.
"Antigen-specific interaction is at the heart of TCR-T cell therapy.
To develop an effective T cell-based therapeutic, it is necessary to discover not only the right TCR, but also the most suitable target epitope."
- Thomas Holberg Blicher, PhD, Senior Research Scientist, Immudex
"MHC Multimers are the gold standard for establishing that something is a real epitope."
- Scientist, Pharmaceutical Company

