MR1 dCODE Dextramer®
For analysis of ligand-specific MAIT cells
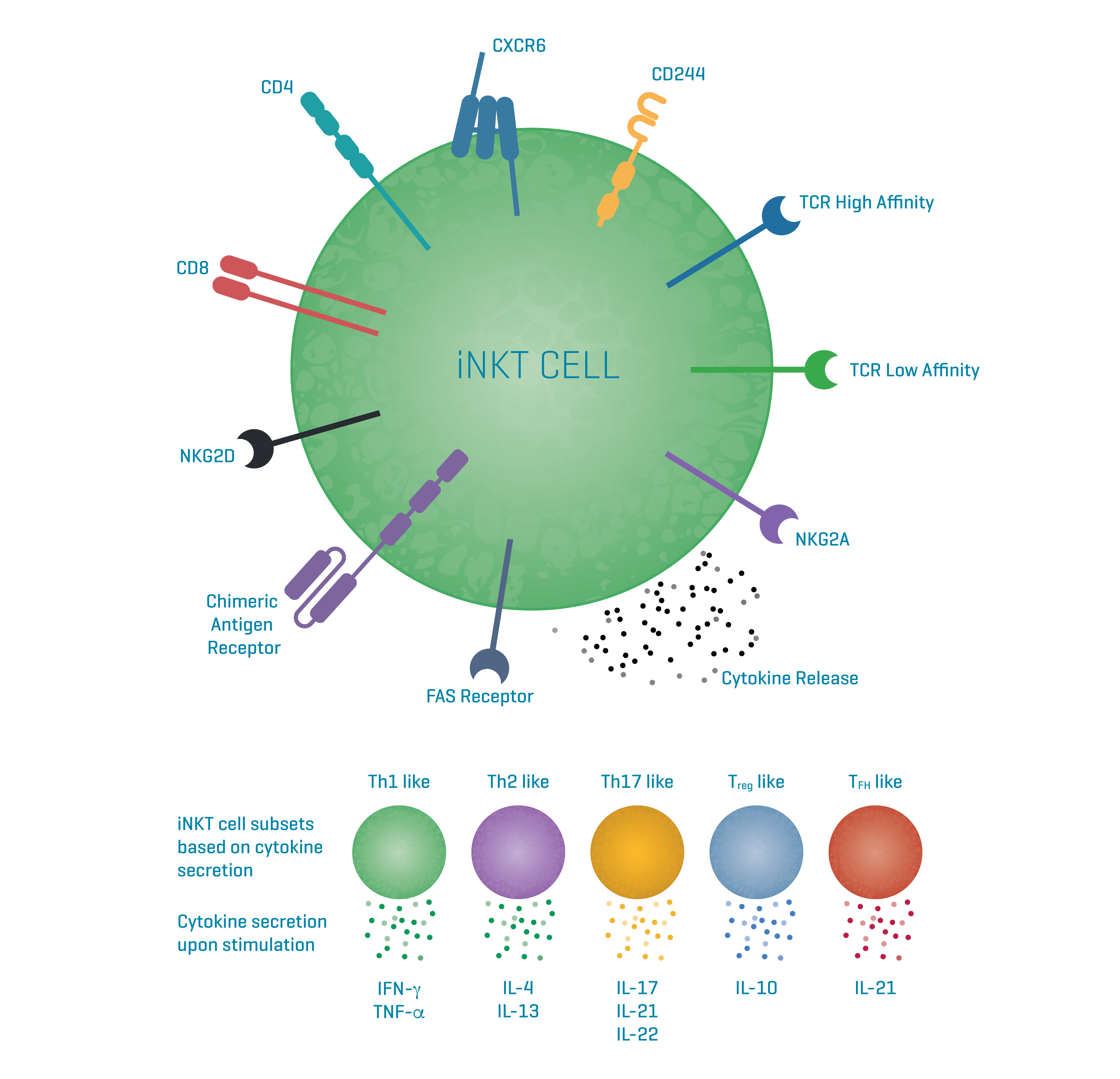
iNKT cell receptors are semi-invariant, primarily featuring an invariant α-chain and a variable β-chain with a diverse CDR3 region. While the β-chain doesn't directly interact with lipid antigens, it influences TCR CD1d-lipid interaction by determining affinity through contacts with the α-chain and CD1d. This likely regulates iNKT cell immune responses to various stimuli.
Current knowledge comes from bulk sequencing of expanded cells, leaving gaps in understanding the relationship between TCR repertoire and tissue type, especially in response to environmental changes. Single-cell multi-omics with DNA-barcoded CD1d dCODE Dextramer® now allows in-depth analysis of iNKT markers, gene expression, and TCR sequences, addressing these questions.
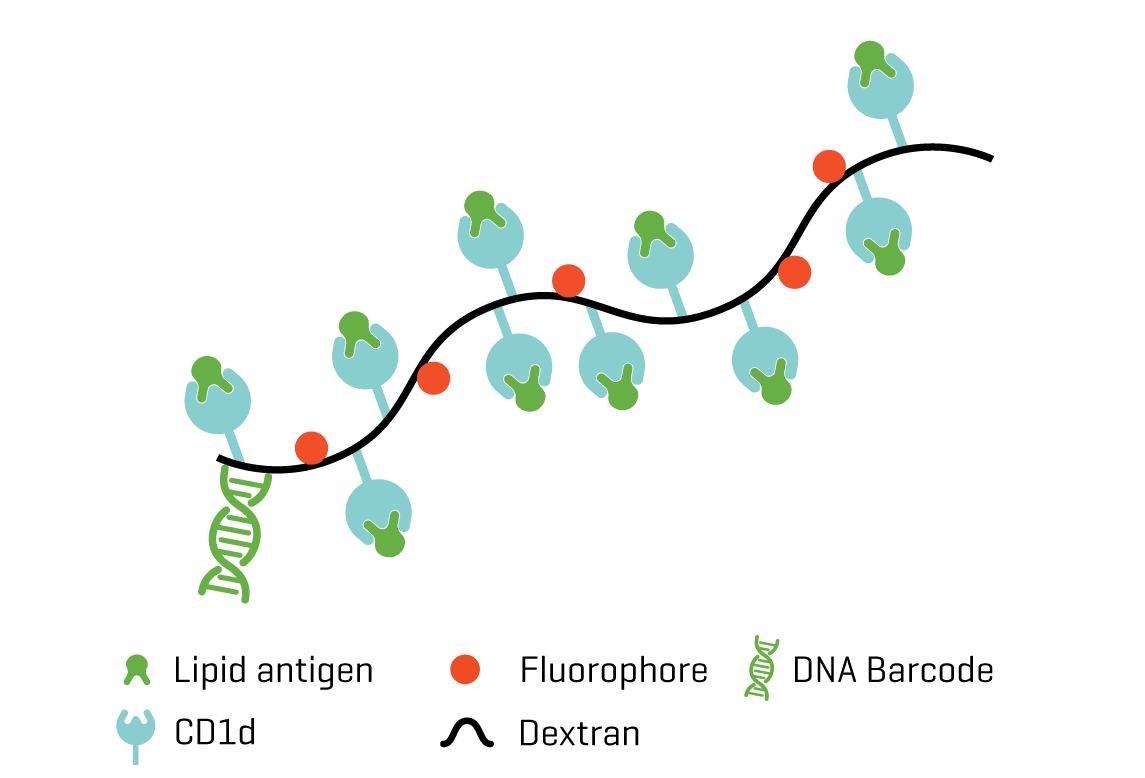
A distinguishing feature of iNKT cells is their reactivity with the lipid α-Galactosylceramide (α-GalCer) when presented by CD1d.
Immudex provides dCODE Dextramer® with CD1d:
DNA barcodes of CD1d dCODE Dextramer® are compatible with 10x Chromium and BD Rhapsody single cell analysis platforms which makes it possible to link TCR sequence, surface marker expression and whole transcriptome analysis in individual CD1d/α-GalCer stained cells.
In a proof-of-concept study we used CD1d/α-GalCer dCODE Dextramer® to sequence the TCR of CD3+ iNKT cells directly from a PBMC sample without prior expansion. The experiment also involved multiplex dCODE Dextramer staining for additional four more cell types (results not reported here).
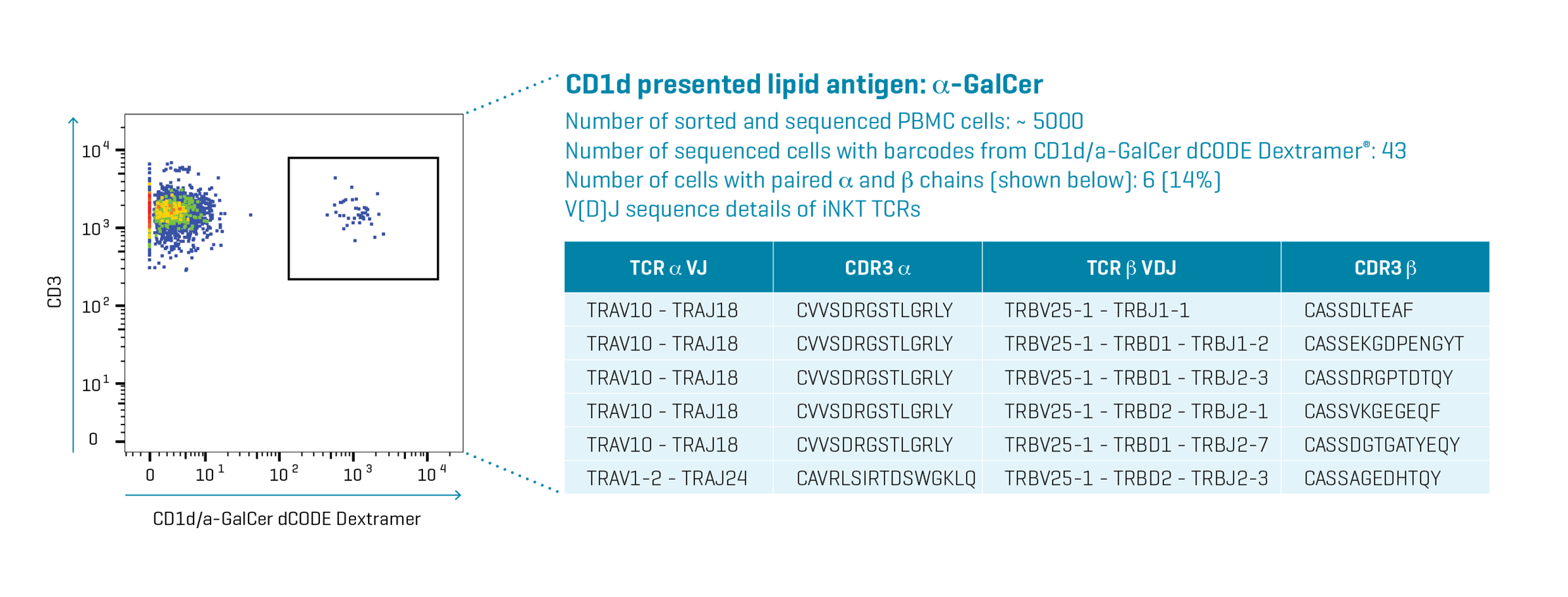
In accordance with previously published results, we found that five out of six paired sequences had an invariable α-VJ region encoded by TRAV10-TRAJ18 segments, with identical CDR3α sequence: VVSDRGSTLGRLY.
All six identified variable β-VDJ regions were encoded by TRBV25-1 and various TRBJ segments and all CDR3β sequences were different.
The α-chain of the last TCR in the list is different. iNKT cells in blood with diverse glycolipid antigen recognition and atypical TCRs have been reported before.
Learn more about compatibility with the BD Rhapsody™ or 10x Chromium platforms.
Immudex provides flexibility for your iNKT cell research by offering biotinylated CD1d Monomers and CD1d Dextramer® and CD1d dCODE Dextramer® reagents for your assay of choice.
To order CD1d products, please send an e-mail to [email protected] specifying information on:
Please note: CD1d dCODE Dextramer® is available as a standard product with the PE fluorochrome, or with BV421, FITC or APC fluorochromes (or no fluorochrome) via Custom Solutions and Services.
|
Cat No |
Product |
CD1d |
Antigen |
Label |
Amount |
|
XD08001XG XD08001RG |
CD1d dCODE Dextramer® (10x) CD1d dCODE Dextramer® (RiO) |
Human CD1d |
Unloaded |
PE |
25 / 50 /150 tests* |
|
XD08002XG XD08002RG |
CD1d dCODE Dextramer® (10x) CD1d dCODE Dextramer® (RiO) |
Human CD1d |
α-GalCer |
PE |
25 / 50 /150 tests* |
|
YD08001XG YD08001RG |
CD1d dCODE Dextramer® (10x) CD1d dCODE Dextramer® (RiO) |
Mouse CD1d |
Unloaded |
PE |
25 / 50 /150 tests* |
|
YD08002XG YD08002RG |
CD1d dCODE Dextramer® (10x) CD1d dCODE Dextramer® (RiO) |
Mouse CD1d |
α-GalCer |
PE |
25 / 50 /150 tests* |
*Higher amounts or test sizes are available upon request.
Do you have questions about the CD1d dCODE Dextramer® technology, implementing our technology into your experimental workflow, protocol optimization, or data analysis and interpretation?
Our technical support team consists of highly skilled scientists that are here to empower you to improve your results, provide you with the tools for success, and share our expertise.
BV421 is equivalent to Brilliant Violet™ 421, which is a trademark or registered trademark of Becton, Dickinson and Company or its affiliates, and is used under license. Powered by BD Innovation.